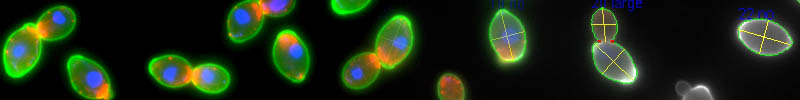
Summary of dataset
Non-essential gene deletion mutants in drug hypersensitive genetic background
The data sheets here are the results created by reanalyzing the images in Ohnuki & Ogawa et al. (2022, NPJ Syst. Biol. Appl.).
- 1982 gene deletion mutants for nonessential genes in drug hypersensitive genetic background
- Average data (16.9 MB)
- Number of cells for ratio parameter (403 KB)
- Number of cells in specimen for ratio parameter (491 MB)
- 749 replicated wild-type (drug-hypersensitive strain)